Modeling the COVID-19 Outbreak with J
Foreword: I am not an epidemiologist and I don’t claim to be. I am, however, a math major and an academic and I feel like I’ve done my due dilligence in reporting this accurately and correctly. The full code used is at the end of the post, if you’d rather not sift through data.
If you’ve been watching the news, I assume you’ve heard about the COVID-19 pandemic. In light of the CDC releasing a model predicting 62% of Americans will become infected1, I became interested in how they came to those numbers. In fact, how do we come to any conclusions as to the spread of disease at all?
There are plenty of well established models for disease spread. The simplest model is the SIR model5, but it comes with a few caveats. The first caveat is that it assumes that once a disease runs its course in a single person, that person will now be permanently immune to the disease. The second caveat is that the model assumes that all infectious people immediately begin to show symptoms. Neither of these assumptions are accurate for COVID-19: the disease has a 14% reinfection rate6, and it takes 10 to 14 days for symptoms to appear.
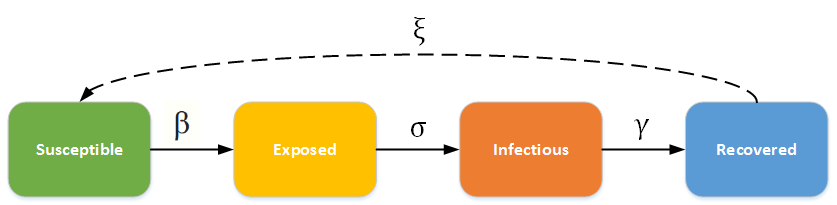
The model that has been used for the current COVID-19 pandemic is the SEIRS model*,4,5. With the SEIR model, we have 4 states that any particular person can be in: “Susceptible”, “Exposed”, “Infectious”, and “Recovered”7.
- A person is susceptible if they are at risk for catching a disease.
- A person is exposed if they have caught the disease or are carrying the disease without any symptoms. This happens during the incubation period.
- A person is infectious if they have now caught the disease and are experiencing symptoms. This is when people go to the hospital, require intensive care, etc.
- A person is recovered if they have gotten over the disease and now have some semblance of immunity.
We have 4 variables we can tweak: \(\beta, \sigma, \gamma, \xi \). They are all rates over a generic unit of time.
- \(\beta\) is the rate at which susceptible people become exposed.
- \(\sigma\) is the rate at which exposed people become infectious.
- \(\gamma\) is the rate at which infectious people recover.
- \(\xi\) is the rate at which recovered people lose immunity and become susceptible.
Used with attribution from https://www.idmod.org/docs/hiv/_images/SEIR-SEIRS.png. Copyright 2019, Intellectual Ventures Management, LLC (IVM). All rights reserved.
These states and variables all have differential equations which correlate them. There are still few notes to be made, though. Firstly, this model doesn’t represent mortality in the population due to the disease. Because of that, the basic reproductive rate4,7 (that is, the amount of people one person will infect, on average) of the disease is quite simple to calculate: \(R_0 = \frac{\beta}{\gamma}\).
We’re going to complicate things a little bit, though. Just looking at disease numbers is boring and unimpactful! Instead of analyzing the differential equations, we’re going to take a look at an actual discrete time simulation which uses those 4 variables. But first, we have to figure out what those variables should be.
Let’s take our “unit of time” to be one day. Here’s some of the information that we have for the virus:
-
We know that \(R_0\) is about 2.288 for COVID-19.
-
We know that it has an incubation period of about 10-ish days (2-14, as per the CDC).
-
We know that once symptoms emerge, they persist about two weeks.9
-
We know that 14%6 of infected people become re-infected.
We can calculate all the variables we need from these, starting with \(\sigma\). Each day, there’s a certain chance \(\sigma\) for COVID-19 to begin showing symptoms. Each day, the chance that it cumulatively hasn’t shown symptoms is \((1 - \sigma)^N\), where \(N\) is the number of days. This is a simple binomial distribution, and for it to begin showing symptoms after 10 days on average means that we must simply solve for \((1 - \sigma)^{10} = 0.5\). So, \(\sigma = 0.066967\)
Once symptoms emerge, they persist about two weeks. Let’s do the same process we did above: \((1 - \gamma)^{14} = 0.5\) means we have \(\gamma = 0.0483048\).
We can calculate \(\beta\) using \(R_0 = \frac{\beta}{\gamma}\) with the \(R_0\) and \(\gamma\) values established above8. \(2.28 = \frac{\beta}{0.0483048}\) means \(\beta = 0.110134944\).
All’s left is to calculate \(\xi\). I have no clue how we could do this, but let’s just say (as a completely random, reasonable guess) that it takes about 30 days to lose immunity to COVID-19. \((1 - \xi)^{30} = 0.5\) gives us \(\xi = 0.02284\).
Now that we have all our variables, let’s boot up our J session and start writing this simulation!
First, numeric definitions.
ppl =: 10 NB. How many people are in our simulation?
Beta =: 0.110134944 NB. The rate at which susceptible people become exposed.
Sigma =: 0.066967 NB. The rate at which exposed people become infectious.
Gamma =: 0.0483048 NB. The rate at which infectious people recover.
Xi =: 0.02284 NB. The rate at which recovered people lose immunity and become susceptible.
R =: 2.28 NB. How many people will 1 person infect?
CFR =: 0.02 NB. Case Fatality Rate
D =: 0.00155285 NB. Death rate per day while symptomatic
Note that we added one extra parameter, the Case Fatality Rate9. This is the percent of COVID-19 cases which end in death. The CDC suggests using 0.25% to 3%, so I’d say 2% is a solid medium, especially with as overwhelmed our US medical systems will be. We can then take that value, and the fact that it takes 13 days to go from symptoms to death9 in the formula \(0.98= (1-D)^{13}\) to get that the probability of death while symptomatic per day is \(D = 0.00155285\)
If we want to simulate the disease’s spread, we have to simulate social contact. So, let’s make a hermitian matrix representing how close people are.
] closeness =: 2 %~ (+ |:) ? (ppl,ppl) $ 0
0.989266 0.433774 0.587923 0.860137 0.423383 0.669393 0.296438 0.658781 0.253612 0.327771
0.433774 0.797844 0.412043 0.734478 0.693995 0.452715 0.585249 0.67874 0.591025 0.897202
0.587923 0.412043 0.916071 0.336036 0.652928 0.531987 0.541966 0.477779 0.522089 0.293315
0.860137 0.734478 0.336036 0.125376 0.438783 0.451785 0.695368 0.468632 0.508739 0.603744
0.423383 0.693995 0.652928 0.438783 0.557975 0.450049 0.722475 0.560327 0.676762 0.809449
0.669393 0.452715 0.531987 0.451785 0.450049 0.435358 0.839259 0.223557 0.835785 0.611344
0.296438 0.585249 0.541966 0.695368 0.722475 0.839259 0.380239 0.36727 0.321142 0.258312
0.658781 0.67874 0.477779 0.468632 0.560327 0.223557 0.36727 0.0571881 0.41249 0.63065
0.253612 0.591025 0.522089 0.508739 0.676762 0.835785 0.321142 0.41249 0.483952 0.657467
0.327771 0.897202 0.293315 0.603744 0.809449 0.611344 0.258312 0.63065 0.657467 0.83782
This matrix represents how likely the \(i\)th and \(j\)th person are to be in social contact with each other. Since it doesn’t make sense for someone to give the virus to themselves, let’s get rid of the middle diagonal.
] risk =: closeness - closeness * =i.ppl
0 0.433774 0.587923 0.860137 0.423383 0.669393 0.296438 0.658781 0.253612 0.327771
0.433774 0 0.412043 0.734478 0.693995 0.452715 0.585249 0.67874 0.591025 0.897202
0.587923 0.412043 0 0.336036 0.652928 0.531987 0.541966 0.477779 0.522089 0.293315
0.860137 0.734478 0.336036 0 0.438783 0.451785 0.695368 0.468632 0.508739 0.603744
0.423383 0.693995 0.652928 0.438783 0 0.450049 0.722475 0.560327 0.676762 0.809449
0.669393 0.452715 0.531987 0.451785 0.450049 0 0.839259 0.223557 0.835785 0.611344
0.296438 0.585249 0.541966 0.695368 0.722475 0.839259 0 0.36727 0.321142 0.258312
0.658781 0.67874 0.477779 0.468632 0.560327 0.223557 0.36727 0 0.41249 0.63065
0.253612 0.591025 0.522089 0.508739 0.676762 0.835785 0.321142 0.41249 0 0.657467
0.327771 0.897202 0.293315 0.603744 0.809449 0.611344 0.258312 0.63065 0.657467 0
Now, let’s make our people! Let 0 represent a dead person, 1 represents a susceptible person, 2 represents an exposed person, 3 represents an infectious person, and finally 4 represents a recovered person.
Our population can then be 10 susceptible people with one infected person.
] starting_pop =: 2 , (ppl - 1) $ 1
2 1 1 1 1 1 1 1 1 1
Let’s then define the infect function which, given a population, advances the disease by one timestep. It’ll be a monadic (one-argument) function of the population:
infect =: 3 : 0
We’re now in the definition for infect. Let’s get a boolean vector representing who’s infectious in the population.
can_spread =. (1&< *. 4&>) y
Let’s use use this vector on the risk matrix, so that each row represents the collective social contact that the nth person in the jth column has with an infected person.
infectiousness =. can_spread ,./ . * risk
Let’s look at what we’ve got at this point (output statements added for clarity).
infect =: 3 : 0
smoutput ] can_spread =. (1&< *. 4&>) y
smoutput ] infectiousness =. can_spread ,./ . * risk
)
starting_pop
2 1 1 1 1 1 1 1 1 1
risk
0 0.320427 0.628024 0.856909 0.705072 0.162919 0.684089 0.0256438 0.531406 0.88479
0.320427 0 0.653424 0.152774 0.607565 0.589595 0.232512 0.534223 0.724831 0.739889
0.628024 0.653424 0 0.485708 0.693178 0.307283 0.455467 0.381945 0.193344 0.431928
0.856909 0.152774 0.485708 0 0.748798 0.0610626 0.490139 0.803612 0.724202 0.437314
0.705072 0.607565 0.693178 0.748798 0 0.658209 0.722431 0.330002 0.355295 0.69828
0.162919 0.589595 0.307283 0.0610626 0.658209 0 0.306342 0.590838 0.779528 0.632452
0.684089 0.232512 0.455467 0.490139 0.722431 0.306342 0 0.820687 0.481649 0.315347
0.0256438 0.534223 0.381945 0.803612 0.330002 0.590838 0.820687 0 0.638455 0.561562
0.531406 0.724831 0.193344 0.724202 0.355295 0.779528 0.481649 0.638455 0 0.770651
0.88479 0.739889 0.431928 0.437314 0.69828 0.632452 0.315347 0.561562 0.770651 0
infect starting_pop
1 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0.320427 0 0 0 0 0 0 0 0 0
0.628024 0 0 0 0 0 0 0 0 0
0.856909 0 0 0 0 0 0 0 0 0
0.705072 0 0 0 0 0 0 0 0 0
0.162919 0 0 0 0 0 0 0 0 0
0.684089 0 0 0 0 0 0 0 0 0
0.0256438 0 0 0 0 0 0 0 0 0
0.531406 0 0 0 0 0 0 0 0 0
0.88479 0 0 0 0 0 0 0 0 0
We see now that infect starting_pop prints that person 1 is infectious, and they’re putting a couple people at very large risk – namely, person 4 with contact level 0.856909 and person 10 with contact level 0.88479.
In our simulation, human contact averages a level of 0.5, so we’ll use that as a baseline. Anything above 0.5 increases \(\beta\), and anything below 0.5 decreases \(\beta\). This simulates reality, with people tending to transfer disease to people they’re closer to more readily.
Let’s now calculate risk for every person in the population seperately. There are 4 risks we’re working with here, and we can represent each of them with vectors of their respective probabilities. Let’s place people into those groups, first:
infect =: 3 : 0
can_spread =. (1&< *. 4&>) y
susceptible =. y = 1
exposed =. y = 2
infectious =. y = 3
recovered =. y = 4
smoutput ] infectiousness =. can_spread ,./ . * risk
)
Now let’s find each person’s level of at risk social contact, and clamp that somehow. This gets a little hand wavy… what happens if all of a person’s sick friends have a closeness rating of 1? Epidemiological models usually don’t need to take social closeness into consideration when simulating populations, because they generally don’t simulate individual people themselves. So, I’m instead going to average all of the values of someone’s social closeness over the amount of possible people that can infect. This will effectively turn someone’s social closeness into a binomial distribution, with a maximum value of 1 and a minimum value of 0 and a mode of 0.5. I’d rather scale this down though, so it has a minimum of -0.1ish, a maximum of 0.1ish, and a mode of 0, so that being social/asocial is not the only defining aspect of whether someone falls ill. This looks like:
5 %~ 0.5 -~ (+/ can_spread) %~ +/ |: susceptible * infectiousness
The rest of the function is simple enough, we just add in the rates for people in each category, then we calculate some random numbers for each person per day and advance them through the illness if they are so unlucky. This is now the full infect function.
infect =: 3 : 0
can_spread =. (1&< *. 4&>) y
susceptible =. y = 1
exposed =. y = 2
infectious =. y = 3
recovered =. y = 4
infectiousness =. can_spread ,./ . * risk
susceptible_to_exposed =. (Beta * susceptible) + 5 %~ 0.5 -~ (+/ can_spread) %~ +/ |: susceptible * infectiousness
exposed_to_infectious =. Sigma * exposed
infectious_to_recovered =. Gamma * infectious
infectious_to_dead =. D * infectious
recovered_to_susceptible =. Xi * recovered
1 (I.recovered_to_susceptible>?ppl$0)} 4 (I.infectious_to_recovered>?ppl$0)} 0 (I.infectious_to_dead>?ppl$0)} 3 (I.exposed_to_infectious>?ppl$0)} 2 (I.susceptible_to_exposed>?ppl$0)} y
)
Finally, let’s make a function which prints out our population.
infect_display =: 3 : 0
out =. infect y
smoutput out
out
)
We can finally simulate our population! Let’s run our simulation for 100 days with 10 people.
starting_pop
2 2 1 1 1 1 1 1 1 1
infect_display^:100 starting_pop
2 2 1 1 2 1 2 1 1 1
2 2 1 1 2 1 3 1 1 1
2 2 1 1 2 1 3 1 1 2
2 2 2 1 2 1 3 1 1 2
3 2 2 1 2 1 3 2 1 2
3 2 2 1 2 1 3 2 2 2
3 2 2 1 2 1 3 2 2 2
3 2 2 1 2 1 3 2 2 2
4 2 2 2 2 1 3 3 2 2
4 3 2 2 2 1 3 3 2 2
4 3 2 2 2 1 3 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 2 2 2 1 4 3 2 2
4 3 3 2 2 1 4 3 2 2
4 3 3 2 2 1 4 3 2 2
4 4 3 2 2 2 4 3 2 2
4 4 3 2 2 2 4 3 2 2
4 4 3 2 2 2 4 3 2 2
4 4 3 2 3 2 4 3 2 2
4 4 3 3 3 2 4 4 2 2
4 4 3 4 3 2 4 4 2 2
4 4 1 4 3 2 4 4 3 2
4 4 1 4 3 2 4 4 3 2
4 4 2 4 3 2 4 4 3 2
4 4 2 4 3 2 4 4 3 2
4 4 2 4 3 2 4 4 3 2
4 4 2 4 3 2 4 4 3 2
4 4 2 4 0 2 4 4 3 2
4 4 2 4 0 2 4 4 3 2
4 4 2 4 0 2 4 4 3 2
4 4 2 4 0 2 4 4 4 2
4 4 2 4 0 2 4 4 4 2
4 4 2 4 0 2 4 4 4 2
4 4 2 4 0 2 4 4 4 2
4 4 2 4 0 2 4 4 4 2
4 4 3 4 0 2 4 4 4 2
4 4 3 4 0 2 4 4 4 2
4 4 3 4 0 2 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 1 4 4 4 3
4 4 4 4 0 1 4 4 4 3
4 4 4 4 0 1 4 4 4 3
4 4 4 4 0 1 4 4 4 3
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 1
4 4 4 4 0 1 4 4 4 2
4 4 4 4 0 2 4 4 4 2
4 4 4 4 0 2 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
4 4 4 4 0 3 4 4 4 2
Uh oh! Looks like 10% of our population passed away. 7 people became immune, 1 person was still infectious after 100 days (seems like they caught it real late), and 1 person was still asymptomatic. Nobody lost their immunity.
Let’s rerun our simulation, this time over 1,000 days with a population of 1,000. This one’ll take a while, the social closeness matrix is now 1,000x1,000 = 1 million entries. We could do some optimizations, like ensuring in-place array transforms and using sparse matricies, but that’s outside of the scope of this blog post.
infect_display^:1000 starting_pop
2 2 1 2 1 1 1 1 1 1 1 1 1 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 2 1 2 1 1 1 1 1 1 2 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 1 1 2 1 2 1 2 1 1 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 2 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 2 ...
2 2 1 2 1 1 1 1 1 1 1 2 1 2 2 2 1 1 1 1 1 1 1 2 1 1 1 1 2 1 2 1 1 2 1 2 1 2 1 1 1 1 2 2 1 1 1 1 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 2 2 1 1 2 1 2 2 2 1 2 2 2 1 1 1 1 1 2 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 2 1 1 2 1 1 1 1 1 2 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 2 ...
3 2 1 2 1 1 1 1 1 1 1 2 1 2 3 2 1 1 1 1 1 1 1 2 1 1 1 1 2 1 2 1 1 2 1 2 1 3 1 1 1 1 2 2 1 1 1 1 2 1 1 1 1 1 1 2 1 1 1 1 2 1 1 2 2 2 1 3 1 3 2 2 1 2 2 2 1 1 1 1 1 2 1 2 2 1 1 1 1 1 2 1 1 1 2 2 1 1 2 1 1 2 1 1 1 1 1 2 2 1 1 1 1 1 1 2 1 1 1 1 1 1 2 1 1 1 1 2 ...
3 2 1 2 1 1 1 1 1 1 1 2 1 2 3 2 1 1 1 1 1 1 1 3 1 1 1 1 2 1 2 1 1 2 1 2 1 3 1 1 1 1 2 2 1 1 1 1 2 1 1 1 2 1 1 2 2 1 2 2 2 1 1 2 2 2 1 3 1 3 2 2 1 2 2 2 1 1 1 1 1 2 1 2 2 1 1 1 1 2 2 1 1 1 2 2 1 1 3 1 1 2 1 1 1 1 1 2 2 1 2 1 1 1 1 2 1 1 1 2 1 1 2 1 2 1 1 2 ...
3 2 2 2 1 1 1 1 1 1 1 2 1 2 3 2 1 2 1 1 1 1 1 3 1 1 1 2 2 1 2 1 1 3 1 2 1 3 1 1 1 1 2 2 1 1 1 1 2 1 1 1 2 1 1 2 2 1 2 2 2 1 1 2 2 2 1 3 1 3 2 2 1 2 2 2 1 1 1 1 1 2 1 2 3 1 1 1 1 2 2 1 1 1 2 2 1 1 3 1 1 2 1 1 1 2 1 2 3 1 2 1 1 1 2 2 1 1 1 3 1 1 2 1 2 1 1 2 ...
3 2 2 2 1 1 1 1 1 1 1 2 1 3 3 2 1 2 1 1 1 1 1 3 1 1 1 2 3 2 3 1 1 3 1 2 2 3 1 1 1 1 2 2 1 2 1 1 2 1 1 1 2 1 1 2 2 1 2 2 2 2 2 2 2 2 2 3 2 3 2 2 1 2 2 2 1 1 1 1 2 3 1 2 4 1 1 1 1 2 2 1 1 1 2 2 1 1 3 1 1 2 1 2 1 2 1 2 3 1 2 1 1 1 2 2 2 1 1 3 1 1 2 1 3 1 2 2 ...
3 2 2 2 1 1 1 1 1 1 1 2 1 3 3 2 1 3 1 1 1 1 1 3 1 1 2 2 3 2 3 1 1 3 1 2 2 3 1 2 1 1 2 2 1 2 1 1 2 1 1 1 2 1 1 2 2 2 2 2 2 2 2 2 2 2 2 3 2 3 2 2 1 2 2 2 1 1 2 1 2 3 1 2 4 2 1 1 1 2 2 1 1 1 2 2 1 2 3 1 1 2 1 2 1 2 1 2 3 1 2 1 1 1 2 2 2 1 1 3 1 1 2 1 4 1 2 2 ...
3 2 2 2 1 1 2 1 1 1 1 2 1 3 3 2 1 3 2 1 1 1 1 3 1 1 2 2 3 2 3 1 1 3 1 2 2 3 1 2 1 1 2 2 1 2 1 1 2 1 1 1 2 1 1 2 2 2 2 2 2 2 2 2 2 2 2 4 2 3 2 2 1 2 2 2 2 1 2 1 2 3 1 3 4 3 2 1 1 2 2 2 2 1 2 2 1 2 3 1 1 2 1 2 1 2 1 2 3 1 2 1 1 1 3 2 2 1 1 3 1 1 2 1 4 2 2 2 ...
3 2 2 2 1 1 2 1 1 1 1 2 1 3 3 2 1 3 2 1 1 1 1 3 1 1 2 2 3 2 3 1 1 3 1 2 2 3 1 2 1 2 2 2 1 2 1 1 2 2 1 1 2 1 2 2 2 2 2 2 2 2 2 2 2 3 2 4 3 3 2 2 1 2 2 2 2 1 2 1 2 3 1 3 4 3 2 1 1 2 2 2 3 1 2 3 1 3 3 1 1 2 1 2 1 2 1 2 3 1 2 1 1 1 3 2 2 1 1 3 1 2 2 1 4 2 2 2 ...
3 2 2 2 1 1 2 1 2 2 1 2 1 3 3 2 1 3 2 1 1 1 1 3 1 1 2 2 3 2 3 2 1 3 1 2 2 3 1 2 2 2 2 2 1 2 1 1 2 2 1 1 2 1 2 2 2 2 2 2 2 2 2 2 2 3 2 4 3 3 2 2 1 2 2 2 2 1 2 1 2 3 1 3 4 3 2 1 1 2 2 3 3 1 2 3 1 3 3 1 1 2 1 2 1 2 2 2 3 1 2 1 1 1 3 2 2 1 1 3 2 2 2 1 4 2 2 2 ...
3 2 3 2 1 1 2 1 2 2 1 2 1 3 3 2 1 3 2 1 1 2 1 3 1 1 3 2 3 2 4 2 1 3 2 2 2 3 1 2 2 2 2 2 1 2 1 1 2 2 1 1 2 1 2 2 2 2 2 2 2 2 2 2 3 3 2 4 3 3 2 2 2 2 3 2 2 1 2 1 2 3 2 3 4 3 2 1 2 2 2 3 3 1 2 3 1 3 4 1 1 2 1 2 1 2 2 2 3 1 2 1 1 1 3 2 2 1 1 3 2 2 2 1 4 3 3 2 ...
3 2 3 2 1 1 2 1 2 2 2 2 1 3 3 2 1 3 2 1 2 2 1 3 1 1 3 2 3 2 4 2 1 3 2 2 2 3 1 2 2 2 2 2 2 2 1 1 2 2 2 1 2 1 2 2 2 2 2 2 2 2 2 2 3 3 2 4 3 3 2 2 2 2 3 2 2 1 2 1 3 3 2 3 4 3 2 1 2 2 2 3 3 1 2 3 1 3 4 1 1 2 1 2 1 2 2 2 3 1 2 1 1 1 3 2 2 1 1 3 2 2 3 1 4 3 3 2 ...
3 2 4 2 1 1 2 1 2 2 2 2 1 3 3 2 1 3 2 2 2 2 2 3 1 1 3 2 3 2 4 2 1 4 2 2 2 3 1 2 2 2 2 2 2 2 1 1 2 2 2 1 2 1 2 2 2 2 2 3 2 2 2 2 3 3 2 4 3 3 2 2 2 2 3 2 2 1 2 1 3 3 2 3 4 3 2 1 3 2 2 3 3 2 2 3 2 3 4 1 1 3 1 2 1 2 2 3 4 1 3 1 2 1 3 3 2 1 1 3 2 2 3 1 4 3 3 3 ...
...snip...
2 3 4 4 3 3 4 0 0 2 0 0 4 4 3 4 4 3 0 4 0 4 0 4 4 4 4 2 2 2 0 4 0 3 0 4 3 0 1 0 4 0 4 3 3 4 4 0 0 4 4 4 4 0 0 0 0 3 2 0 4 1 3 0 4 4 3 0 2 0 0 4 1 3 0 4 4 4 3 0 2 4 2 4 2 0 3 4 3 4 0 4 4 2 2 3 0 2 3 4 4 0 4 4 0 4 3 4 0 4 2 0 0 4 0 1 3 0 1 3 4 4 4 0 4 0 4 0 ...
2 3 4 4 3 3 4 0 0 2 0 0 4 4 3 4 4 3 0 4 0 4 0 4 4 4 4 2 2 2 0 4 0 3 0 4 3 0 2 0 4 0 4 3 3 4 4 0 0 4 4 4 4 0 0 0 0 3 2 0 4 1 3 0 4 4 3 0 2 0 0 4 2 3 0 4 4 4 3 0 2 4 2 4 2 0 3 4 3 4 0 4 4 2 2 4 0 2 3 4 4 0 4 4 0 4 3 4 0 4 2 0 0 4 0 1 3 0 1 3 4 4 4 0 4 0 4 0 ...
2 3 4 4 3 3 4 0 0 2 0 0 4 4 3 4 4 3 0 4 0 4 0 4 4 4 4 2 2 2 0 4 0 3 0 4 3 0 2 0 4 0 4 3 3 4 4 0 0 4 4 4 4 0 0 0 0 3 2 0 4 1 3 0 4 4 3 0 2 0 0 4 2 3 0 4 4 4 3 0 2 4 2 4 2 0 3 4 3 4 0 4 4 2 2 4 0 2 3 4 4 0 4 4 0 4 3 4 0 4 2 0 0 4 0 1 3 0 1 3 4 4 4 0 4 0 4 0 ...
Remember, every 0 is a single dead person. In our simulations, a closed population that doesn’t enact any sort of social distancing will suffer a massive death toll. Let’s make a little table of the various populations and rerun our simulation, this time over the course of a year.
infect_display =: 3 : 0
out =. infect y
smoutput out
smoutput ('Susceptible';'Exposed';'Infectious';'Recovered';'Dead'),:(# 1=out);(# 2=out);(# 3=out);(# 4=out);(# 0=out)
out
)
infect_display^:365 starting_pop
2 2 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 2 1 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 2 1 2 1 2 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│886 │114 │0 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 2 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 2 1 2 1 1 1 1 2 1 2 1 1 1 1 1 1 1 2 2 2 1 2 2 1 1 1 1 1 1 2 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 2 1 1 1 1 1 2 1 2 2 1 1 1 2 1 1 1 1 2 1 1 1 1 1 1 2 1 1 1 2 1 1 1 1 1 2 1 1 1 1 1 2 1 2 2 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│780 │207 │13 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 3 1 1 1 1 1 1 1 1 3 1 1 1 2 1 1 2 1 1 1 2 1 1 2 1 1 1 2 1 2 1 2 1 2 1 1 2 1 2 1 1 1 1 1 1 1 2 2 2 2 2 2 1 1 2 1 1 2 3 1 1 1 1 1 2 1 2 2 1 1 1 1 1 1 1 2 2 1 1 1 2 1 2 1 2 2 1 1 1 2 2 1 1 1 2 1 1 2 2 1 1 2 1 1 1 2 1 1 1 1 1 2 1 1 1 1 1 2 1 2 2 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│693 │279 │28 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 3 1 2 1 1 1 1 2 1 3 1 1 1 2 1 1 2 1 1 1 2 1 1 2 1 1 1 2 2 2 1 2 1 2 1 1 2 1 2 1 1 2 1 1 2 1 2 2 2 2 2 2 1 1 2 1 1 2 3 1 1 1 1 1 2 1 2 2 1 2 1 1 1 1 2 2 2 1 1 1 2 1 2 1 2 3 1 1 1 2 2 1 1 1 3 1 1 2 2 1 1 2 1 1 1 2 1 1 2 1 1 2 1 1 2 1 2 2 1 2 2 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│619 │334 │45 │2 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
...
After one day, we already see 114 people exposed to the disease. Therein lies the danger of COVID-19 – it incubates in the body, undetected. This number is a bit of an exaggeration, as this population is closed (nobody enters and exits), and everyone interacts with everyone else every day. If Edgar Allen Poe knew about COVID-19, these are the parameters under which “The Masque of the Red Death” was written. So, just keep it in mind that these numbers don’t entirely represent how the disease will spread in places less crowded than, say, Times Square.
Let’s now skip to the end of the year.
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│101 │163 │178 │443 │115 │
└───────────┴───────┴──────────┴─────────┴────┘
4 2 4 3 3 3 3 3 2 4 4 0 1 0 2 2 4 0 3 4 4 4 1 4 4 3 3 4 2 0 2 4 4 0 2 0 3 2 4 2 2 4 3 2 4 4 4 4 3 4 4 4 3 0 3 2 4 3 0 4 3 4 0 1 0 4 0 2 4 4 1 3 0 2 3 3 3 3 1 0 4 2 3 2 2 4 4 1 0 0 1 3 2 3 1 4 4 4 4 0 4 0 4 2 0 4 2 4 1 0 3 4 0 3 4 4 2 1 4 3 4 2 0 0 4 1 4 3 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│96 │165 │189 │435 │115 │
└───────────┴───────┴──────────┴─────────┴────┘
4 2 4 3 3 3 3 3 2 4 4 0 2 0 2 2 4 0 3 4 4 4 1 4 4 3 3 4 2 0 2 4 4 0 2 0 3 2 4 2 3 4 3 2 4 4 4 4 3 4 4 4 3 0 4 2 4 3 0 4 3 4 0 1 0 4 0 2 4 4 2 3 0 2 3 3 3 3 2 0 4 2 3 2 2 4 4 2 0 0 1 3 2 3 1 4 4 4 4 0 4 0 4 2 0 4 2 4 1 0 3 4 0 3 4 4 2 1 4 3 4 2 0 0 4 1 4 3 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│89 │164 │196 │436 │115 │
└───────────┴───────┴──────────┴─────────┴────┘
4 2 4 3 3 3 3 3 2 4 4 0 2 0 2 2 4 0 3 4 4 4 1 4 4 3 3 4 2 0 2 4 4 0 2 0 3 2 4 2 3 4 3 2 4 4 4 4 3 4 4 4 3 0 4 3 4 3 0 4 3 4 0 1 0 4 0 2 4 4 2 3 0 2 4 3 3 3 2 0 4 2 3 2 2 4 4 2 0 0 1 3 3 3 1 4 4 4 4 0 4 0 4 2 0 4 2 4 1 0 3 4 0 3 4 4 2 1 4 3 4 2 0 0 4 1 4 3 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│81 │163 │198 │443 │115 │
└───────────┴───────┴──────────┴─────────┴────┘
4 2 4 3 3 3 3 3 2 4 4 0 2 0 2 2 4 0 3 4 4 1 1 4 4 3 3 4 2 0 2 4 4 0 2 0 3 2 4 2 3 4 3 2 4 4 4 4 3 4 4 4 3 0 4 3 4 3 0 4 3 4 0 1 0 4 0 2 4 4 2 3 0 2 4 3 3 3 2 0 4 2 3 2 2 4 4 2 0 0 2 3 3 4 1 4 4 4 4 0 4 0 4 2 0 4 2 4 1 0 3 4 0 3 4 4 2 1 4 4 4 2 0 0 4 1 4 3 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│81 │163 │196 │445 │115 │
└───────────┴───────┴──────────┴─────────┴────┘
115 dead. Let’s enact some social distancing and give the entire population the asociality modifier of -0.1.
risk =: (ppl,ppl)$0
infect_display^:365 starting_pop
2 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│987 │13 │0 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│977 │23 │0 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│968 │31 │1 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
2 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 1 1 1 2 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 2 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│955 │42 │3 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
...snip...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│499 │84 │103 │251 │63 │
└───────────┴───────┴──────────┴─────────┴────┘
1 4 1 3 3 1 1 4 4 1 1 3 4 1 0 1 4 1 2 1 2 1 1 0 1 1 1 1 1 4 1 3 3 3 3 1 1 3 1 1 4 1 3 4 4 1 1 1 1 0 1 4 4 1 1 1 0 4 4 1 1 4 2 1 1 1 1 1 4 1 1 1 3 2 1 4 0 4 4 1 1 1 4 1 2 4 4 3 4 4 4 1 4 0 1 3 2 2 4 4 1 1 2 4 4 1 2 2 1 1 1 1 4 2 3 1 1 1 2 1 4 1 1 3 4 2 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│497 │86 │104 │250 │63 │
└───────────┴───────┴──────────┴─────────┴────┘
1 4 1 3 3 1 1 4 4 1 1 3 4 1 0 1 4 1 2 1 3 1 1 0 1 2 1 1 1 4 1 3 3 3 3 1 1 3 1 1 4 1 3 4 4 1 1 1 1 0 1 4 4 1 1 1 0 4 4 1 1 4 2 1 1 1 1 1 4 1 1 1 3 2 1 4 0 4 4 1 1 1 4 1 2 4 4 3 4 4 4 1 4 0 1 3 2 2 4 4 1 1 2 4 4 1 2 2 1 1 1 1 4 2 3 1 1 1 2 1 4 1 1 4 4 2 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│500 │83 │105 │248 │64 │
└───────────┴───────┴──────────┴─────────┴────┘
1 4 1 3 3 1 1 4 4 1 1 3 1 1 0 1 4 1 2 1 3 1 1 0 1 3 1 1 1 4 1 3 3 3 3 2 1 3 1 1 4 1 3 4 4 1 1 1 1 0 1 4 4 1 1 1 0 4 1 1 1 4 2 1 1 1 1 1 4 1 1 1 3 2 1 4 0 4 4 1 1 1 4 1 2 4 4 3 4 4 4 1 4 0 1 3 2 2 4 4 1 1 2 4 4 1 2 2 1 1 1 1 4 2 3 1 1 1 2 1 4 1 1 1 4 2 2 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│500 │89 │104 │243 │64 │
└───────────┴───────┴──────────┴─────────┴────┘
1 4 1 3 3 1 1 4 4 1 1 3 1 1 0 1 4 1 2 1 3 1 1 0 1 3 1 1 1 4 1 3 3 3 3 2 1 3 1 1 4 1 3 4 4 1 1 1 1 0 1 4 4 1 1 1 0 4 1 1 1 4 2 1 1 1 1 1 4 1 1 1 3 2 1 4 0 4 4 1 1 1 4 1 2 4 4 3 4 4 4 1 4 0 1 3 2 2 4 4 1 1 2 4 4 1 2 2 1 1 1 1 4 2 3 1 1 1 2 1 4 1 1 1 4 2 2 1 ...
Just by coming together as a society to cut down on the spread of COVID-19, without changing anything else, we can halve the death toll by the end of the first year according to the model. That number, of course, relies on the assumptions we made on how we calculate social-ness. Let’s try calculating it in a different way – instead of being a flat additive ±0.1 modifier on \(\beta\), how about we make it multiplicative?
A COVID-19 superspreader has infected up to 11 other people10, which is about 5x what \(R\) suggests. This suggests that the standard deviation of human relationships probably hovers around \(\frac{5}{3}\), putting superspreaders in the top 0.3% of human contact if contact is normally distributed.
Our social risk matrix is now generated thusly:
ClosenessStdDev =: 5 % 3
ClosenessMean =: 1
rand_norm =: 3 : '(2&o. 2p1 * ? y) * %: _2 * ^. ? y'
closeness =: 2 %~ (+ |:) ClosenessMean + ClosenessStdDev * rand_norm (ppl,ppl) $ 0
risk =: closeness - closeness * =i.ppl
And it produces risk matricies like this:
risk
0 1.79563 0.7175 2.94724 2.02043 0.192731 1.91119 0.919267 1.44283 0.703641
1.79563 0 2.5158 0.312935 _0.268321 1.52827 1.8885 _0.529835 0.636972 0.368671
0.7175 2.5158 0 _0.00583566 _1.69494 3.41282 0.539451 3.15925 2.79567 0.749702
2.94724 0.312935 _0.00583566 0 1.80479 0.805175 1.01254 2.35566 1.63717 0.0811308
2.02043 _0.268321 _1.69494 1.80479 0 _0.176758 0.678648 1.02585 1.6886 3.44681
0.192731 1.52827 3.41282 0.805175 _0.176758 0 0.905996 2.21435 1.01778 1.96473
1.91119 1.8885 0.539451 1.01254 0.678648 0.905996 0 _0.285226 2.50235 2.85363
0.919267 _0.529835 3.15925 2.35566 1.02585 2.21435 _0.285226 0 _1.20884 _0.310217
1.44283 0.636972 2.79567 1.63717 1.6886 1.01778 2.50235 _1.20884 0 2.53888
0.703641 0.368671 0.749702 0.0811308 3.44681 1.96473 2.85363 _0.310217 2.53888 0
The exposure risk is now mutiplicative:
susceptible_to_exposed =. (Beta * susceptible) * (+/ can_spread) %~ +/ |: susceptible * infectiousness
And produces results like this (starting conditions same as before, 1000 people, 2 exposed, 365 days):
infect_display^:365 starting_pop
...snip...
4 1 1 4 3 4 2 4 4 3 4 4 4 3 4 4 3 3 2 2 4 4 3 1 2 4 2 4 0 4 3 2 2 2 3 1 4 4 4 4 4 4 4 4 4 4 4 4 4 2 4 3 4 0 0 4 2 3 4 4 4 4 0 3 4 0 0 2 2 4 4 4 4 0 4 4 4 3 2 0 3 2 0 4 0 4 2 3 1 2 4 4 0 0 0 3 4 1 3 4 1 4 3 4 0 1 2 2 2 4 4 1 1 0 0 4 4 0 1 4 3 3 0 4 4 1 4 0 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│86 │150 │192 │431 │141 │
└───────────┴───────┴──────────┴─────────┴────┘
As our social contact is now multiplicative with \(\beta\), we can now accurately answer questions like “What if people were half as likely to have social contact?” by dividing the distribution in half. With all parameters the same, but with half as much multiplicative social contact, the death toll has gone down by a third:
infect_display^:365 starting_pop
...snip...
2 4 4 4 1 1 4 2 2 3 1 1 4 0 4 4 1 4 2 2 2 4 4 1 4 4 3 0 3 4 4 2 4 4 1 3 1 4 3 3 4 4 1 0 4 1 1 2 3 4 2 3 3 0 4 3 4 4 2 4 1 0 2 0 4 3 1 4 3 2 4 2 4 1 4 4 4 0 1 2 4 4 2 4 3 0 1 4 3 3 4 4 1 4 4 3 4 0 1 2 4 3 2 4 3 4 3 4 4 1 4 4 3 4 0 0 1 4 4 2 4 2 1 4 3 4 4 4 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│168 │136 │173 │422 │101 │
└───────────┴───────┴──────────┴─────────┴────┘
What if we cut our social contact down to one quarter? This is what I’ve been trying to do in real life – I usually go out with people almost every day, and I have been limiting that now to once or twice per week. What if our whole society did that?**
infect_display^:365 starting_pop
...snip...
1 2 2 4 4 2 1 4 3 2 2 1 3 2 4 1 3 4 3 3 1 1 4 4 0 1 4 3 1 3 0 1 1 1 1 4 4 3 2 1 0 4 3 4 1 1 1 4 3 4 4 3 4 0 4 3 2 1 1 1 1 0 4 0 4 0 4 1 0 1 0 3 4 3 3 2 4 2 2 1 3 1 4 1 4 1 1 1 3 1 1 3 4 0 4 1 3 1 3 4 1 1 1 1 1 1 1 4 2 1 2 4 4 1 2 3 0 2 2 4 4 2 4 1 4 2 2 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│299 │117 │172 │320 │92 │
└───────────┴───────┴──────────┴─────────┴────┘
Just for fun, what would happen if nobody came into contact with each other at all anymore?
infect_display^:100 starting_pop
2 3 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│998 │1 │1 │0 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
...snip...
4 3 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│998 │0 │1 │1 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
4 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│998 │0 │0 │2 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
4 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...
┌───────────┬───────┬──────────┬─────────┬────┐
│Susceptible│Exposed│Infectious│Recovered│Dead│
├───────────┼───────┼──────────┼─────────┼────┤
│998 │0 │0 │2 │0 │
└───────────┴───────┴──────────┴─────────┴────┘
Unsurprisingly, COVID-19 dies out after 24 days.
Food for thought. If you liked this, or found this helpful, please throw a coffee my way at https://paypal.me/tslimkemann.
Sources:
-
https://thehill.com/homenews/state-watch/487489-worst-case-coronavirus-models-show-massive-us-toll
-
https://www.cdc.gov/csels/dsepd/ss1978/lesson3/section3.html
-
https://en.wikipedia.org/wiki/Compartmental_models_in_epidemiology
-
https://www.newsobserver.com/news/nation-world/national/article241209786.html
Footnotes:
*: The basic SEIRS model is not actually the model used for COVID-19. The model used is much closer to the one presented in Li, Fuxiang, and Xiao-Qiang Zhao. “A Periodic SEIRS Epidemic Model with a Time-dependent Latent Period.” Journal of Mathematical Biology 78.5 (2019): 1553-579. Web.
**: This model suggests some serious seasonality to COVID-19. After analyzing all the data, the change here is so little in this simulation because it actually strikes in two waves – once initially, and once again once immunity wears off after three months. Note the much higher number of susceptible people and the much lower number of recovered people.
Full code:
ppl =: 1000 NB. How many people are in our simulation?
Beta =: 0.110134944 NB. The rate at which susceptible people become exposed.
Sigma =: 0.066967 NB. The rate at which exposed people become infectious.
Gamma =: 0.0483048 NB. The rate at which infectious people recover.
Xi =: 0.02284 NB. The rate at which recovered people lose immunity and become susceptible.
R =: 2.28 NB. How many people will 1 person infect?
CFR =: 0.02 NB. Case Fatality Rate
D =: 0.00155285 NB. Death rate per day while symptomatic
ClosenessStdDev =: 5 % 3
ClosenessMean =: 1
rand_norm =: 3 : '(2&o. 2p1 * ? y) * %: _2 * ^. ? y'
closeness =: 4 %~ 2 %~ (+ |:) ClosenessMean + ClosenessStdDev * rand_norm (ppl,ppl) $ 0
risk =: closeness - closeness * =i.ppl
NB. risk =: (ppl,ppl)$0
NB. 0 = Dead
NB. 1 = Susceptible
NB. 2 = Exposed
NB. 3 = Infectious
NB. 4 = Recovered
starting_pop =: 2 2 , (ppl - 2) $ 1
infect =: 3 : 0
can_spread =. (1&< *. 4&>) y
susceptible =. y = 1
exposed =. y = 2
infectious =. y = 3
recovered =. y = 4
infectiousness =. can_spread ,./ . * risk
susceptible_to_exposed =. (Beta * susceptible) * (+/ can_spread) %~ +/ |: susceptible * infectiousness
exposed_to_infectious =. Sigma * exposed
infectious_to_recovered =. Gamma * infectious
infectious_to_dead =. D * infectious
recovered_to_susceptible =. Xi * recovered
1 (I.recovered_to_susceptible>?ppl$0)} 4 (I.infectious_to_recovered>?ppl$0)} 0 (I.infectious_to_dead>?ppl$0)} 3 (I.exposed_to_infectious>?ppl$0)} 2 (I.susceptible_to_exposed>?ppl$0)} y
)
infect_display =: 3 : 0
out =. infect y
smoutput out
smoutput ('Susceptible';'Exposed';'Infectious';'Recovered';'Dead'),:(+/1=out);(+/2=out);(+/3=out);(+/4=out);(+/0=out)
out
)